gstools.covmodel.base¶
GStools subpackage providing the base class for covariance models.
The following classes are provided
CovModel([dim, var, len_scale, nugget, …]) |
Base class for the GSTools covariance models |
-
class
gstools.covmodel.base.CovModel(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
objectBase class for the GSTools covariance models
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}. Default:None
Examples
>>> from gstools import CovModel >>> import numpy as np >>> class Gau(CovModel): ... def correlation(self, r): ... return np.exp(-(r/self.len_scale)**2) ... >>> model = Gau() >>> model.spectrum(2) 0.00825830126008459
Attributes: anglesnumpy.ndarray: The rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parametersdimint: The dimension of the model.dist_funchankel_kwdict: Keywords forhankel.SymmetricFourierTransformhas_cdfbool: State if a cdf is defined by the userhas_ppfbool: State if a ppf is defined by the userintegral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scalelen_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel classnuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nuggetopt_argopt_arg_boundsdict: Bounds for the optional argumentssillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variancevar_rawfloat: The raw variance of the model without factor
Methods
check_arg_bounds()Here the arguments are checked to be within the given bounds check_opt_arg()Here you can run checks for the optional arguments default_arg_bounds()Here you can provide a dictionary with default boundaries for the standard arguments. default_opt_arg()Here you can provide a dictionary with default values for the optional arguments. default_opt_arg_bounds()Here you can provide a dictionary with default boundaries for the optional arguments. fit_variogram(x_data, y_data, **para_deselect)fit the isotropic variogram-model to given data fix_dim()Set a fix dimension for the model ln_spectral_rad_pdf(r)The log radial spectral density of the model depending on the dimension percentile_scale([per])calculate the distance, where the given percentile of the variance is reached by the variogram set_arg_bounds(**kwargs)Set bounds for the parameters of the model spectral_density(k)The spectral density of the covariance model. spectral_rad_pdf(r)The radial spectral density of the model depending on the dimension spectrum(k)The spectrum of the covariance model. var_factor()Optional factor for the variance -
check_opt_arg()[source]¶ Here you can run checks for the optional arguments
This is in addition to the bound-checks
Notes
- You can use this to raise a ValueError/warning
- Any return value will be ignored
- This method will only be run once, when the class is initialized
-
default_arg_bounds()[source]¶ Here you can provide a dictionary with default boundaries for the standard arguments.
-
default_opt_arg()[source]¶ Here you can provide a dictionary with default values for the optional arguments.
-
default_opt_arg_bounds()[source]¶ Here you can provide a dictionary with default boundaries for the optional arguments.
-
fit_variogram(x_data, y_data, **para_deselect)[source]¶ fit the isotropic variogram-model to given data
Parameters: - x_data (
numpy.ndarray) – The radii of the meassured variogram. - y_data (
numpy.ndarray) – The messured variogram - **para_deselect – You can deselect the parameters to be fitted, by setting them “False” as keywords. By default, all parameters are fitted.
Returns: - fit_para (
dict) – Dictonary with the fitted parameter values - pcov (
numpy.ndarray) – The estimated covariance of popt fromscipy.optimize.curve_fit
Notes
You can set the bounds for each parameter by accessing
CovModel.set_arg_bounds.The fitted parameters will be instantly set in the model.
- x_data (
-
ln_spectral_rad_pdf(r)[source]¶ The log radial spectral density of the model depending on the dimension
-
percentile_scale(per=0.9)[source]¶ calculate the distance, where the given percentile of the variance is reached by the variogram
-
set_arg_bounds(**kwargs)[source]¶ Set bounds for the parameters of the model
Parameters: **kwargs – Parameter name as keyword (“var”, “len_scale”, “nugget”, <opt_arg>) and a list of 2 or 3 values as value:
[a, b]or[a, b, <type>]
<type> is one of
"oo","cc","oc"or"co"to define if the bounds are open (“o”) or closed (“c”).
-
spectral_density(k)[source]¶ The spectral density of the covariance model.
This is given by:
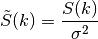
Where
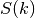 is the spectrum of the covariance model.
is the spectrum of the covariance model.Parameters: k ( float) – Radius of the phase: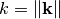
-
spectrum(k)[source]¶ The spectrum of the covariance model.
This is given by:
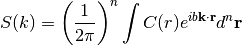
Internally, this is calculated by the hankel transformation:
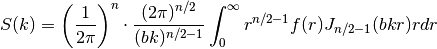
Where
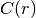 is the covariance function of the model.
is the covariance function of the model.Parameters: k ( float) – Radius of the phase: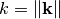
-
angles¶ The rotation angles (in rad) of the model.
Type: numpy.ndarray
-
anis¶ The anisotropy factors of the model.
Type: numpy.ndarray
-
arg_bounds¶ Bounds for all parameters
Notes
Keys are the opt-arg names and values are lists of 2 or 3 values:
[a, b]or[a, b, <type>]
<type> is one of
"oo","cc","oc"or"co"to define if the bounds are open (“o”) or closed (“c”).Type: dict
-
hankel_kw¶ Keywords for
hankel.SymmetricFourierTransformType: dict
-
integral_scale¶ The main integral scale of the model.
Raises: ValueError– If integral scale is not setable.Type: float
-
integral_scale_vec¶ The integral scales in each direction.
Notes
- This is calculated by:
integral_scale_vec[0] = integral_scaleintegral_scale_vec[1] = integral_scale*anis[0]integral_scale_vec[2] = integral_scale*anis[1]
Type: numpy.ndarray
-
len_scale_bounds¶ Bounds for the lenght scale
Notes
Is a list of 2 or 3 values:
[a, b]or[a, b, <type>]
<type> is one of
"oo","cc","oc"or"co"to define if the bounds are open (“o”) or closed (“c”).Type: list
-
len_scale_vec¶ The length scales in each direction.
Notes
- This is calculated by:
len_scale_vec[0] = len_scalelen_scale_vec[1] = len_scale*anis[0]len_scale_vec[2] = len_scale*anis[1]
Type: numpy.ndarray
-
nugget_bounds¶ Bounds for the nugget
Notes
Is a list of 2 or 3 values:
[a, b]or[a, b, <type>]
<type> is one of
"oo","cc","oc"or"co"to define if the bounds are open (“o”) or closed (“c”).Type: list
-
opt_arg_bounds¶ Bounds for the optional arguments
Notes
Keys are the opt-arg names and values are lists of 2 or 3 values:
[a, b]or[a, b, <type>]
<type> is one of
"oo","cc","oc"or"co"to define if the bounds are open (“o”) or closed (“c”).Type: dict
- dim (