gstools.covmodel.models¶
GStools subpackage providing different covariance models.
The following classes and functions are provided
Gaussian([dim, var, len_scale, nugget, …]) |
The Gaussian covariance model. |
Exponential([dim, var, len_scale, nugget, …]) |
The Exponential covariance model. |
Matern([dim, var, len_scale, nugget, anis, …]) |
The Matérn covariance model. |
Stable([dim, var, len_scale, nugget, anis, …]) |
The stable covariance model. |
Rational([dim, var, len_scale, nugget, …]) |
The rational quadratic covariance model. |
Linear([dim, var, len_scale, nugget, anis, …]) |
The bounded linear covariance model. |
Circular([dim, var, len_scale, nugget, …]) |
The circular covariance model. |
Spherical([dim, var, len_scale, nugget, …]) |
The Spherical covariance model. |
Intersection([dim, var, len_scale, nugget, …]) |
The Intersection covariance model. |
-
class
gstools.covmodel.models.Gaussian(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe Gaussian covariance model.
Notes
This model is given by the following correlation function:
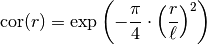
.
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Gaussian correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Provide default optional arguments by the user. default_opt_arg_bounds()Provide default boundaries for optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_cdf(r)Radial spectral cdf. spectral_rad_pdf(r)Radial spectral density of the model. spectral_rad_ppf(u)Radial spectral ppf. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
covariance(r)¶ Covariance of the model.
Given by:
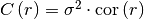
Where
 is the correlation function.
is the correlation function.
-
spectral_density(k)[source]¶ Spectral density of the covariance model.
This is given by:
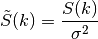
Where
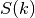 is the spectrum of the covariance model.
is the spectrum of the covariance model.Parameters: k ( float) – Radius of the phase: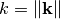
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
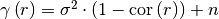
Where
 is the correlation function.
is the correlation function.
- dim (
-
class
gstools.covmodel.models.Exponential(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe Exponential covariance model.
Notes
This model is given by the following correlation function:
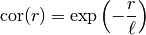
.
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Exponential correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Provide default optional arguments by the user. default_opt_arg_bounds()Provide default boundaries for optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_cdf(r)Radial spectral cdf. spectral_rad_pdf(r)Radial spectral density of the model. spectral_rad_ppf(u)Radial spectral ppf. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
covariance(r)¶ Covariance of the model.
Given by:
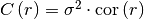
Where
 is the correlation function.
is the correlation function.
-
spectral_density(k)[source]¶ Spectral density of the covariance model.
This is given by:
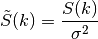
Where
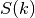 is the spectrum of the covariance model.
is the spectrum of the covariance model.Parameters: k ( float) – Radius of the phase: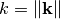
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
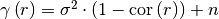
Where
 is the correlation function.
is the correlation function.
- dim (
-
class
gstools.covmodel.models.Matern(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe Matérn covariance model.
Notes
This model is given by the following correlation function:
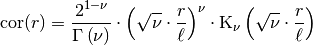
Where
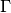 is the gamma function and
is the gamma function and 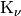 is the modified Bessel function of the second kind.
is the modified Bessel function of the second kind.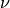 is a shape parameter and should be >= 0.2.
is a shape parameter and should be >= 0.2.If
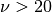 , a gaussian model is used, since it is the limit
case:
, a gaussian model is used, since it is the limit
case: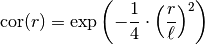
Other Parameters: - **opt_arg – The following parameters are covered by these keyword arguments
- nu (
float, optional) – Shape parameter. Standard range:[0.2, 30]Default:1.0 - .
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Matérn correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Defaults for the optional arguments. default_opt_arg_bounds()Defaults for boundaries of the optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
covariance(r)¶ Covariance of the model.
Given by:
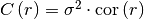
Where
 is the correlation function.
is the correlation function.
-
default_opt_arg()[source]¶ Defaults for the optional arguments.
{"nu": 1.0}
Returns: Defaults for optional arguments Return type: dict
-
default_opt_arg_bounds()[source]¶ Defaults for boundaries of the optional arguments.
{"nu": [0.5, 30.0, "cc"]}
Returns: Boundaries for optional arguments Return type: dict
-
spectral_density(k)[source]¶ Spectral density of the covariance model.
This is given by:
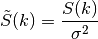
Where
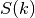 is the spectrum of the covariance model.
is the spectrum of the covariance model.Parameters: k ( float) – Radius of the phase: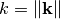
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
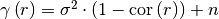
Where
 is the correlation function.
is the correlation function.
-
class
gstools.covmodel.models.Stable(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe stable covariance model.
Notes
This model is given by the following correlation function:
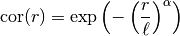
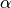 is a shape parameter with
is a shape parameter with ![\alpha\in(0,2]](_images/math/afa9c7c6eefaf7893d2d327325a2fcaf27818e30.png)
Other Parameters: - **opt_arg – The following parameters are covered by these keyword arguments
- alpha (
float, optional) – Shape parameter. Standard range:(0, 2]Default:1.5 - .
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Check the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Stable correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Defaults for the optional arguments. default_opt_arg_bounds()Defaults for boundaries of the optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
check_opt_arg()[source]¶ Check the optional arguments.
Warns: alpha – If alpha is < 0.3, the model tends to a nugget model and gets numerically unstable.
-
covariance(r)¶ Covariance of the model.
Given by:
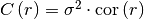
Where
 is the correlation function.
is the correlation function.
-
default_opt_arg()[source]¶ Defaults for the optional arguments.
{"alpha": 1.5}
Returns: Defaults for optional arguments Return type: dict
-
default_opt_arg_bounds()[source]¶ Defaults for boundaries of the optional arguments.
{"alpha": [0, 2, "oc"]}
Returns: Boundaries for optional arguments Return type: dict
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
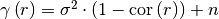
Where
 is the correlation function.
is the correlation function.
-
class
gstools.covmodel.models.Rational(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe rational quadratic covariance model.
Notes
This model is given by the following correlation function:
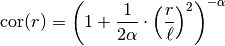
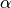 is a shape parameter and should be > 0.5.
is a shape parameter and should be > 0.5.Other Parameters: - **opt_arg – The following parameters are covered by these keyword arguments
- alpha (
float, optional) – Shape parameter. Standard range:(0, inf)Default:1.0 - .
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Rational correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Defaults for the optional arguments. default_opt_arg_bounds()Defaults for boundaries of the optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
covariance(r)¶ Covariance of the model.
Given by:
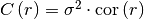
Where
 is the correlation function.
is the correlation function.
-
default_opt_arg()[source]¶ Defaults for the optional arguments.
{"alpha": 1.0}
Returns: Defaults for optional arguments Return type: dict
-
default_opt_arg_bounds()[source]¶ Defaults for boundaries of the optional arguments.
{"alpha": [0.5, inf]}
Returns: Boundaries for optional arguments Return type: dict
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
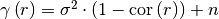
Where
 is the correlation function.
is the correlation function.
-
class
gstools.covmodel.models.Linear(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe bounded linear covariance model.
This model is derived from the relative intersection area of two lines in 1D, where the middle points have a distance of
 and the line lengths are
and the line lengths are 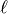 .
.Notes
This model is given by the following correlation function:
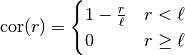
.
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Linear correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Provide default optional arguments by the user. default_opt_arg_bounds()Provide default boundaries for optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
covariance(r)¶ Covariance of the model.
Given by:
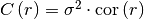
Where
 is the correlation function.
is the correlation function.
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
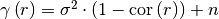
Where
 is the correlation function.
is the correlation function.
- dim (
-
class
gstools.covmodel.models.Circular(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe circular covariance model.
This model is derived as the relative intersection area of two discs in 2D, where the middle points have a distance of
 and the diameters are given by
and the diameters are given by 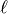 .
.Notes
This model is given by the following correlation function:
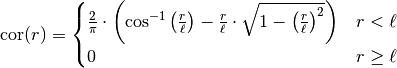
.
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Circular correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Provide default optional arguments by the user. default_opt_arg_bounds()Provide default boundaries for optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
covariance(r)¶ Covariance of the model.
Given by:
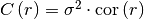
Where
 is the correlation function.
is the correlation function.
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
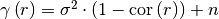
Where
 is the correlation function.
is the correlation function.
- dim (
-
class
gstools.covmodel.models.Spherical(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe Spherical covariance model.
This model is derived from the relative intersection area of two spheres in 3D, where the middle points have a distance of
 and the diameters are given by
and the diameters are given by 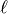 .
.Notes
This model is given by the following correlation function:
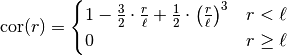
.
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Spherical correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Provide default optional arguments by the user. default_opt_arg_bounds()Provide default boundaries for optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
covariance(r)¶ Covariance of the model.
Given by:
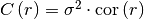
Where
 is the correlation function.
is the correlation function.
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
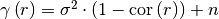
Where
 is the correlation function.
is the correlation function.
- dim (
-
class
gstools.covmodel.models.Intersection(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelThe Intersection covariance model.
This model is derived from the relative intersection area of two d-dimensional spheres, where the middle points have a distance of
 and the diameters are given by
and the diameters are given by 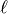 .
.In 1D this is the Linear model, in 2D this is the Circular model and in 3D this is the Spherical model.
Notes
This model is given by the following correlation functions.
In 1D:
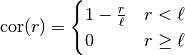
In 2D:
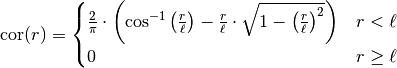
In 3D:
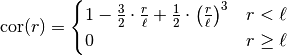
.
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Provide default optional arguments by the user. default_opt_arg_bounds()Provide default boundaries for optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for the variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. correlation -
covariance(r)¶ Covariance of the model.
Given by:
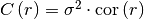
Where
 is the correlation function.
is the correlation function.
-
spectral_density(k)[source]¶ Spectral density of the covariance model.
This is given by:
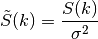
Where
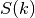 is the spectrum of the covariance model.
is the spectrum of the covariance model.Parameters: k ( float) – Radius of the phase: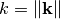
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
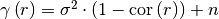
Where
 is the correlation function.
is the correlation function.
- dim (