gstools.covmodel.tpl_models¶
GStools subpackage providing truncated power law covariance models.
The following classes and functions are provided
TPLGaussian([dim, var, len_scale, nugget, …]) |
Truncated-Power-Law with Gaussian modes. |
TPLExponential([dim, var, len_scale, …]) |
Truncated-Power-Law with Exponential modes. |
TPLStable([dim, var, len_scale, nugget, …]) |
Truncated-Power-Law with Stable modes. |
-
class
gstools.covmodel.tpl_models.TPLGaussian(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelTruncated-Power-Law with Gaussian modes.
Notes
The truncated power law is given by a superposition of scale-dependent variograms:
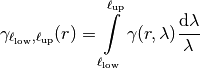
with Gaussian modes on each scale:
![\gamma(r,\lambda) &=
\sigma^2(\lambda)\cdot\left(1-
\exp\left[- \left(\frac{r}{\lambda}\right)^{2}\right]
\right)\\
\sigma^2(\lambda) &= C\cdot\lambda^{2H}](_images/math/e69be21611709aa4654df4c8c978881bb00452c8.png)
This results in:
![\gamma_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}}(r) &=
\sigma^2_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}}\cdot\left(1-
H \cdot
\frac{\ell_{\mathrm{up}}^{2H} \cdot
E_{1+H}
\left[\left(\frac{r}{\ell_{\mathrm{up}}}\right)^{2}\right]
- \ell_{\mathrm{low}}^{2H} \cdot
E_{1+H}
\left[\left(\frac{r}{\ell_{\mathrm{low}}}\right)^{2}\right]}
{\ell_{\mathrm{up}}^{2H}-\ell_{\mathrm{low}}^{2H}}
\right) \\
\sigma^2_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}} &=
\frac{C\cdot\left(\ell_{\mathrm{up}}^{2H}
-\ell_{\mathrm{low}}^{2H}\right)}{2H}](_images/math/d02a2e97cad41efd5e233d579461b9b0ae22d9e5.png)
The “length scale” of this model is equivalent by the integration range:
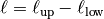
If you want to define an upper scale truncation, you should set
len_lowandlen_scaleaccordingly.The following Parameters occure:
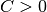 : The scaling factor from the Power-Law.
This parameter will be calculated internally by the given variance.
You can access C directly by
: The scaling factor from the Power-Law.
This parameter will be calculated internally by the given variance.
You can access C directly by model.var_raw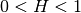 : The hurst coefficient (
: The hurst coefficient (model.hurst)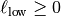 : The lower length scale truncation
of the model (
: The lower length scale truncation
of the model (model.len_low)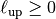 : The upper length scale truncation
of the model (
: The upper length scale truncation
of the model (model.len_up)This will be calculated internally by:
len_up = len_low + len_scale
That means, that the
len_scalein this model actually represents the integration range for the truncated power law.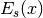 is the exponential integral.
is the exponential integral.
Other Parameters: Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.len_upfloat: Upper length scale truncation of the model.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Truncated-Power-Law with Gaussian modes - correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Defaults for the optional arguments. default_opt_arg_bounds()Defaults for boundaries of the optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for C (Power-Law factor) to result in variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
correlation(r)[source]¶ Truncated-Power-Law with Gaussian modes - correlation function.
If
len_low=0we have a simple representation:![\mathrm{cor}(r) =
H \cdot
E_{1+H}
\left[
\left(\frac{r}{\ell}\right)^{2}
\right]](_images/math/3c3ab271606f69b087812f1eb0f9a9e3432b6270.png)
The general case:
![\mathrm{cor}(r) =
H \cdot
\frac{\ell_{\mathrm{up}}^{2H} \cdot
E_{1+H}
\left[\left(\frac{r}{\ell_{\mathrm{up}}}\right)^{2}\right]
- \ell_{\mathrm{low}}^{2H} \cdot
E_{1+H}
\left[\left(\frac{r}{\ell_{\mathrm{low}}}\right)^{2}\right]}
{\ell_{\mathrm{up}}^{2H}-\ell_{\mathrm{low}}^{2H}}](_images/math/c52cb97e6e4be1d1cb4b01d87e1fef04d399884b.png)
-
covariance(r)¶ Covariance of the model.
Given by:
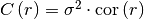
Where
 is the correlation function.
is the correlation function.
-
default_opt_arg()[source]¶ Defaults for the optional arguments.
{"hurst": 0.5, "len_low": 0.0}
Returns: Defaults for optional arguments Return type: dict
-
default_opt_arg_bounds()[source]¶ Defaults for boundaries of the optional arguments.
{"hurst": [0, 1, "oo"], "len_low": [0, 1000, "cc"]}
Returns: Boundaries for optional arguments Return type: dict
-
var_factor()[source]¶ Factor for C (Power-Law factor) to result in variance.
This is used to result in the right variance, which is depending on the hurst coefficient and the length-scale extents
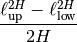
Returns: factor Return type: float
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
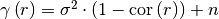
Where
 is the correlation function.
is the correlation function.
-
class
gstools.covmodel.tpl_models.TPLExponential(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelTruncated-Power-Law with Exponential modes.
Notes
The truncated power law is given by a superposition of scale-dependent variograms:
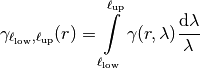
with Exponential modes on each scale:
![\gamma(r,\lambda) &=
\sigma^2(\lambda)\cdot\left(1-
\exp\left[- \frac{r}{\lambda}\right]
\right)\\
\sigma^2(\lambda) &= C\cdot\lambda^{2H}](_images/math/5fe8d8cd20f2aa111aa0bb622a9b0f64248d4190.png)
This results in:
![\gamma_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}}(r) &=
\sigma^2_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}}\cdot\left(1-
2H \cdot
\frac{\ell_{\mathrm{up}}^{2H} \cdot
E_{1+2H}\left[\frac{r}{\ell_{\mathrm{up}}}\right]
- \ell_{\mathrm{low}}^{2H} \cdot
E_{1+2H}\left[\frac{r}{\ell_{\mathrm{low}}}\right]}
{\ell_{\mathrm{up}}^{2H}-\ell_{\mathrm{low}}^{2H}}
\right) \\
\sigma^2_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}} &=
\frac{C\cdot\left(\ell_{\mathrm{up}}^{2H}
-\ell_{\mathrm{low}}^{2H}\right)}{2H}](_images/math/4f939baf88106807c8f4efcca090e76cc86d09fc.png)
The “length scale” of this model is equivalent by the integration range:
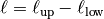
If you want to define an upper scale truncation, you should set
len_lowandlen_scaleaccordingly.The following Parameters occure:
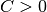 : The scaling factor from the Power-Law.
This parameter will be calculated internally by the given variance.
You can access C directly by
: The scaling factor from the Power-Law.
This parameter will be calculated internally by the given variance.
You can access C directly by model.var_raw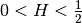 : The hurst coefficient (
: The hurst coefficient (model.hurst)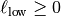 : The lower length scale truncation
of the model (
: The lower length scale truncation
of the model (model.len_low)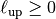 : The upper length scale truncation
of the model (
: The upper length scale truncation
of the model (model.len_up)This will be calculated internally by:
len_up = len_low + len_scale
That means, that the
len_scalein this model actually represents the integration range for the truncated power law.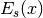 is the exponential integral.
is the exponential integral.
Other Parameters: Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.len_upfloat: Upper length scale truncation of the model.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Run checks for the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Truncated-Power-Law with Exponential modes - correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Defaults for the optional arguments. default_opt_arg_bounds()Defaults for boundaries of the optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for C (Power-Law factor) to result in variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
correlation(r)[source]¶ Truncated-Power-Law with Exponential modes - correlation function.
If
len_low=0we have a simple representation:![\mathrm{cor}(r) =
H \cdot
E_{1+H}
\left[
\frac{r}{\ell}
\right]](_images/math/a2b897ca48935afaf0dcc1619524023496ef2b8e.png)
The general case:
![\mathrm{cor}(r) =
2H \cdot
\frac{\ell_{\mathrm{up}}^{2H} \cdot
E_{1+2H}\left[\frac{r}{\ell_{\mathrm{up}}}\right]
- \ell_{\mathrm{low}}^{2H} \cdot
E_{1+2H}\left[\frac{r}{\ell_{\mathrm{low}}}\right]}
{\ell_{\mathrm{up}}^{2H}-\ell_{\mathrm{low}}^{2H}}](_images/math/f0e761987c8a1ce72a807d06af9312ec2bacb294.png)
-
covariance(r)¶ Covariance of the model.
Given by:
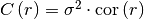
Where
 is the correlation function.
is the correlation function.
-
default_opt_arg()[source]¶ Defaults for the optional arguments.
{"hurst": 0.25, "len_low": 0.0}
Returns: Defaults for optional arguments Return type: dict
-
default_opt_arg_bounds()[source]¶ Defaults for boundaries of the optional arguments.
{"hurst": [0, 1, "oo"], "len_low": [0, 1000, "cc"]}
Returns: Boundaries for optional arguments Return type: dict
-
var_factor()[source]¶ Factor for C (Power-Law factor) to result in variance.
This is used to result in the right variance, which is depending on the hurst coefficient and the length-scale extents
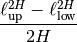
Returns: factor Return type: float
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
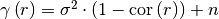
Where
 is the correlation function.
is the correlation function.
-
class
gstools.covmodel.tpl_models.TPLStable(dim=3, var=1.0, len_scale=1.0, nugget=0.0, anis=1.0, angles=0.0, integral_scale=None, var_raw=None, hankel_kw=None, **opt_arg)[source]¶ Bases:
gstools.covmodel.base.CovModelTruncated-Power-Law with Stable modes.
Notes
The truncated power law is given by a superposition of scale-dependent variograms:
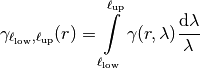
with Stable modes on each scale:
![\gamma(r,\lambda) &=
\sigma^2(\lambda)\cdot\left(1-
\exp\left[- \left(\frac{r}{\lambda}\right)^{\alpha}\right]
\right)\\
\sigma^2(\lambda) &= C\cdot\lambda^{2H}](_images/math/411c0961a08eb64bbff8f97954392676b2b12e82.png)
This results in:
![\gamma_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}}(r) &=
\sigma^2_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}}\cdot\left(1-
\frac{2H}{\alpha} \cdot
\frac{\ell_{\mathrm{up}}^{2H} \cdot
E_{1+\frac{2H}{\alpha}}
\left[\left(\frac{r}{\ell_{\mathrm{up}}}\right)^{\alpha}\right]
- \ell_{\mathrm{low}}^{2H} \cdot
E_{1+\frac{2H}{\alpha}}
\left[\left(\frac{r}{\ell_{\mathrm{low}}}\right)^{\alpha}\right]}
{\ell_{\mathrm{up}}^{2H}-\ell_{\mathrm{low}}^{2H}}
\right) \\
\sigma^2_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}} &=
\frac{C\cdot\left(\ell_{\mathrm{up}}^{2H}
-\ell_{\mathrm{low}}^{2H}\right)}{2H}](_images/math/6c9fcb42a0a2b965650ad0d6a2e9d5f26a462ebf.png)
The “length scale” of this model is equivalent by the integration range:
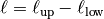
If you want to define an upper scale truncation, you should set
len_lowandlen_scaleaccordingly.The following Parameters occure:
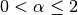 : The shape parameter of the Stable model.
: The shape parameter of the Stable model.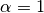 : Exponential modes
: Exponential modes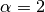 : Gaussian modes
: Gaussian modes
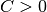 : The scaling factor from the Power-Law.
This parameter will be calculated internally by the given variance.
You can access C directly by
: The scaling factor from the Power-Law.
This parameter will be calculated internally by the given variance.
You can access C directly by model.var_raw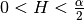 : The hurst coefficient (
: The hurst coefficient (model.hurst)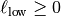 : The lower length scale truncation
of the model (
: The lower length scale truncation
of the model (model.len_low)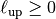 : The upper length scale truncation
of the model (
: The upper length scale truncation
of the model (model.len_up)This will be calculated internally by:
len_up = len_low + len_scale
That means, that the
len_scalein this model actually represents the integration range for the truncated power law.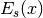 is the exponential integral.
is the exponential integral.
Other Parameters: - **opt_arg – The following parameters are covered by these keyword arguments
- hurst (
float, optional) – Hurst coefficient of the power law. Standard range:(0, 1). Default:0.5 - alpha (
float, optional) – Shape parameter of the stable model. Standard range:(0, 2]. Default:1.5 - len_low (
float, optional) – The lower length scale truncation of the model. Standard range:[0, 1000]. Default:0.0 - .
Parameters: - dim (
int, optional) – dimension of the model. Default:3 - var (
float, optional) – variance of the model (the nugget is not included in “this” variance) Default:1.0 - len_scale (
floatorlist, optional) – length scale of the model. If a single value is given, the same length-scale will be used for every direction. If multiple values (for main and transversal directions) are given, anis will be recalculated accordingly. Default:1.0 - nugget (
float, optional) – nugget of the model. Default:0.0 - anis (
floatorlist, optional) – anisotropy ratios in the transversal directions [y, z]. Default:1.0 - angles (
floatorlist, optional) –angles of rotation:
- in 2D: given as rotation around z-axis
- in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
Default:
0.0 - integral_scale (
floatorlistorNone, optional) – If given,len_scalewill be ignored and recalculated, so that the integral scale of the model matches the given one. Default:None - var_raw (
floatorNone, optional) – raw variance of the model which will be multiplied withCovModel.var_factorto result in the actual variance. If given,varwill be ignored. (This is just for models that overrideCovModel.var_factor) Default:None - hankel_kw (
dictorNone, optional) – Modify the init-arguments ofhankel.SymmetricFourierTransformused for the spectrum calculation. Use with caution (Better: Don’t!).Noneis equivalent to{"a": -1, "b": 1, "N": 1000, "h": 0.001}.
Attributes: anglesnumpy.ndarray: Rotation angles (in rad) of the model.anisnumpy.ndarray: The anisotropy factors of the model.argarg_boundsdict: Bounds for all parameters.dimint: The dimension of the model.dist_funcdo_rotationbool: State if a rotation is performed.hankel_kwdict:hankel.SymmetricFourierTransformkwargs.has_cdfbool: State if a cdf is defined by the user.has_ppfbool: State if a ppf is defined by the user.integral_scalefloat: The main integral scale of the model.integral_scale_vecnumpy.ndarray: The integral scales in each direction.len_scalefloat: The main length scale of the model.len_scale_boundslist: Bounds for the lenght scale.len_scale_vecnumpy.ndarray: The length scales in each direction.len_upfloat: Upper length scale truncation of the model.namestr: The name of the CovModel class.nuggetfloat: The nugget of the model.nugget_boundslist: Bounds for the nugget.opt_argopt_arg_boundsdict: Bounds for the optional arguments.pykrige_angle2D rotation angle for pykrige.
pykrige_angle_x3D rotation angle around x for pykrige.
pykrige_angle_y3D rotation angle around y for pykrige.
pykrige_angle_z3D rotation angle around z for pykrige.
pykrige_anis2D anisotropy ratio for pykrige.
pykrige_anis_y3D anisotropy ratio in y direction for pykrige.
pykrige_anis_z3D anisotropy ratio in z direction for pykrige.
pykrige_kwargsKeyword arguments for pykrige routines.
sillfloat: The sill of the variogram.varfloat: The variance of the model.var_boundslist: Bounds for the variance.var_rawfloat: The raw variance of the model without factor.
Methods
calc_integral_scale()Calculate the integral scale of the isotrope model. check_arg_bounds()Check arguments to be within the given bounds. check_opt_arg()Check the optional arguments. cor_spatial(pos)Spatial correlation respecting anisotropy and rotation. correlation(r)Truncated-Power-Law with Stable modes - correlation function. cov_nugget(r)Covariance of the model respecting the nugget at r=0. cov_spatial(pos)Spatial covariance respecting anisotropy and rotation. covariance(r)Covariance of the model. default_arg_bounds()Provide default boundaries for arguments. default_opt_arg()Defaults for the optional arguments. default_opt_arg_bounds()Defaults for boundaries of the optional arguments. fit_variogram(x_data, y_data[, maxfev])Fiting the isotropic variogram-model to given data. fix_dim()Set a fix dimension for the model. ln_spectral_rad_pdf(r)Log radial spectral density of the model. percentile_scale([per])Calculate the percentile scale of the isotrope model. plot([func])Plot a function of a the CovModel. pykrige_vario([args, r])Isotropic variogram of the model for pykrige. set_arg_bounds(**kwargs)Set bounds for the parameters of the model. spectral_density(k)Spectral density of the covariance model. spectral_rad_pdf(r)Radial spectral density of the model. spectrum(k)Spectrum of the covariance model. var_factor()Factor for C (Power-Law factor) to result in variance. vario_nugget(r)Isotropic variogram of the model respecting the nugget at r=0. vario_spatial(pos)Spatial variogram respecting anisotropy and rotation. variogram(r)Isotropic variogram of the model. -
check_opt_arg()[source]¶ Check the optional arguments.
Warns: alpha – If alpha is < 0.3, the model tends to a nugget model and gets numerically unstable.
-
correlation(r)[source]¶ Truncated-Power-Law with Stable modes - correlation function.
If
len_low=0we have a simple representation:![\mathrm{cor}(r) =
\frac{2H}{\alpha} \cdot
E_{1+\frac{2H}{\alpha}}
\left[
\left(\frac{r}{\ell}\right)^{\alpha}
\right]](_images/math/8677ebcea73d8029050ba3abc94a1495142ae115.png)
The general case:
![\mathrm{cor}(r) =
\frac{2H}{\alpha} \cdot
\frac{\ell_{\mathrm{up}}^{2H} \cdot
E_{1+\frac{2H}{\alpha}}
\left[\left(\frac{r}{\ell_{\mathrm{up}}}\right)^{\alpha}\right]
- \ell_{\mathrm{low}}^{2H} \cdot
E_{1+\frac{2H}{\alpha}}
\left[\left(\frac{r}{\ell_{\mathrm{low}}}\right)^{\alpha}\right]}
{\ell_{\mathrm{up}}^{2H}-\ell_{\mathrm{low}}^{2H}}](_images/math/926f2d4093919024d47f31d0b2ee2ec79c48b2f9.png)
-
covariance(r)¶ Covariance of the model.
Given by:
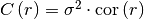
Where
 is the correlation function.
is the correlation function.
-
default_opt_arg()[source]¶ Defaults for the optional arguments.
{"hurst": 0.5, "alpha": 1.5, "len_low": 0.0}
Returns: Defaults for optional arguments Return type: dict
-
default_opt_arg_bounds()[source]¶ Defaults for boundaries of the optional arguments.
{"hurst": [0, 1, "oo"], "alpha": [0, 2, "oc"], "len_low": [0, 1000, "cc"]}
Returns: Boundaries for optional arguments Return type: dict
-
var_factor()[source]¶ Factor for C (Power-Law factor) to result in variance.
This is used to result in the right variance, which is depending on the hurst coefficient and the length-scale extents
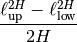
Returns: factor Return type: float
-
variogram(r)¶ Isotropic variogram of the model.
Given by:
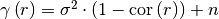
Where
 is the correlation function.
is the correlation function.